Our group works at the interface between chemistry and biology. It is at this interface where many new discoveries about how living systems work are waiting to be uncovered. Our lab works on molecular strategies for uncovering host-pathogen interactions that include genomic and proteomic profiling, activity-based protein profiling, omics studies of microRNAs (miRNAs) and other non-coding RNAs, small molecule probes, bioorthogonal reactions, organisms with expanded genetic codes, and protein-based tools for sensing and imaging of molecular interactions including the viral suppressor of RNA silencing protein p19. We study pathogenesis as well as the immunometabolic response to infection and translate new knowledge into new diagnostic and therapeutic strategies to combat infection as well as developing new biotechnological tools with industrial applications.
|
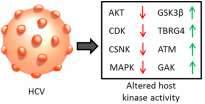
Host-Virus Interactions
Viruses remodel their host cell, targeting host cell pathways to alter the host machinery they use to enter, replicate, assemble, and secrete, and significantly altering the metabolic profile of the cell in the process.
|
ABPPActivity-based protein profiling (ABPP) uses small molecule activity-based probes (ABPs) which form irreversible bonds with target enzymes based on their structure and their catalytic activity.
|
Tools for RNA Silencing
One of our focuses is on the development and use of screening tools, including one’s based on the viral suppressor of RNA silencing protein p19 and small molecule annotation of miRNA function, for the investigation of the roles of small RNAs in disease.
|
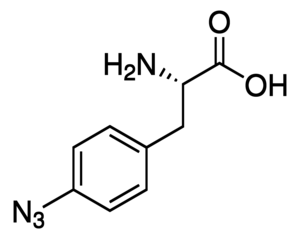
Bioorthogonal chemistry
Our research group explores the field of bioorthogonal chemistry in several significant ways. Focus has been placed on developing and applying novel chemistry for bioorthogonal labelling of different biomaterials.
|
Expanded genetic code technologies
Unnatural amino acid incorporation into proteins allows for new customizable functionality. Our group takes advantage of this tool to study protein-protein interactions and to create tools for probing complex pathways.
|
We kindly thank these funding agencies for generously supporting our research