Our lab employs genetic code expansion (GCE) technology, a powerful protein engineering approach that enables site-specific incorporation of unnatural amino acids (UAAs) into proteins. We apply this strategy to study a range of nucleic acid-binding proteins, allowing us to interrogate RNA- and DNA-protein interactions with exceptional molecular precision. These engineered systems provide unique tools for understanding the roles of such proteins in the context of viral infection and host-pathogen interactions.
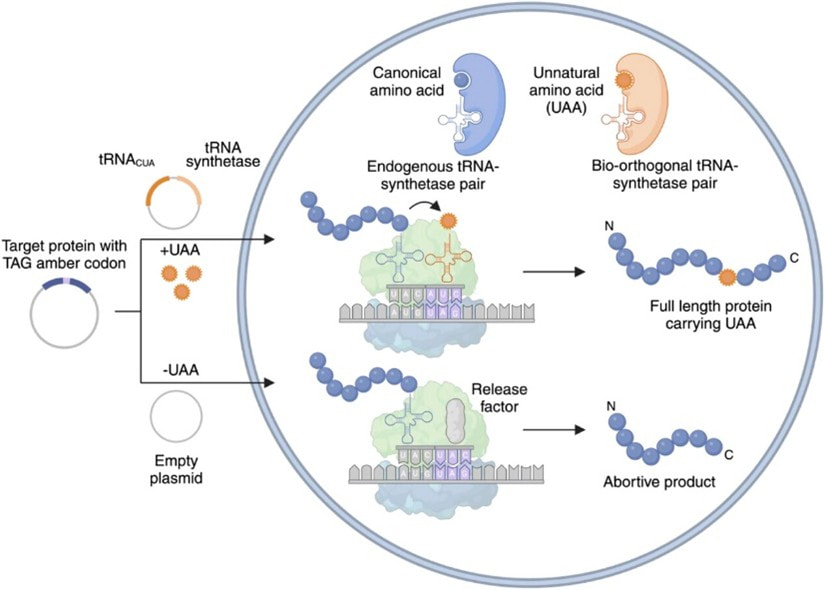
GCE utilizes unnatural amino acids (UAAs) beyond the 20 known canonical amino acids by using engineered, orthogonal tRNA/tRNA synthetase pairs. These pairs are evolved in the laboratory to selectively recognize and incorporate a desired UAA in response to a rare codon. GCE is inherently biorthogonal and introduces minimal interruptions to the system’s endogenous cellular processes.
GCE utilizes unnatural amino acids (UAAs) beyond the 20 known canonical amino acids by using engineered, orthogonal tRNA/tRNA synthetase pairs. These pairs are evolved in the laboratory to selectively recognize and incorporate a desired UAA in response to a rare codon. GCE is inherently biorthogonal and introduces minimal interruptions to the system’s endogenous cellular processes.
Using this technology, our lab has engineered viral RNA silencing suppressor protein, p19, to enable detection of its small non-coding RNA ligands. Site specific incorporation of p-azidophenylalanine allowed functionalization via biorthogonal click chemistry, resulting in a Förster resonance energy transfer (FRET)-based probe. This enabled sensitive detection of RNA binding and delivery of RNA cargo into cells.
We have also shown tested the use of p-azidophenylalanine in protein-protein interaction mapping using novel crosslinking methodologies. With the ability to catch transient protein-protein interactions within the cellular environment, novel cellular pathways and interactions could be discovered and tested.
Our genetic code expansion projects intersect with our interest in helicases in projects involving engineered hepatitis C virus nonstructural protein 3 helicase (NS3h) and SARS-CoV-2 helicase (Nsp13) based-FRET probes to study it helicase function. We utilized genetic code expansion technology to engineer a protein that allows for smFRET experiments. The engineered probe allowed us to monitor the dynamics of enzyme translocation during unwinding in real-time.
We have also shown tested the use of p-azidophenylalanine in protein-protein interaction mapping using novel crosslinking methodologies. With the ability to catch transient protein-protein interactions within the cellular environment, novel cellular pathways and interactions could be discovered and tested.
Our genetic code expansion projects intersect with our interest in helicases in projects involving engineered hepatitis C virus nonstructural protein 3 helicase (NS3h) and SARS-CoV-2 helicase (Nsp13) based-FRET probes to study it helicase function. We utilized genetic code expansion technology to engineer a protein that allows for smFRET experiments. The engineered probe allowed us to monitor the dynamics of enzyme translocation during unwinding in real-time.
Using UAAs with metal-chelating activity, we are engineering entirely new functions into viral helicases and RNA-binding proteins, repurposing them for new cellular activities beyond their typical roles. Building on our previous work, we have incorporated the noncanonical amino acid (2,2-bipyridin-5-yl)alanine (BpyAla) into the viral suppressor protein P19, thereby introducing a tunable metal-dependent RNA-binding and cleavage activity. Leveraging these findings, we now aim to transfer this chemically encoded functionality to additional proteins, including helicases, to create RNA-targeting enzymes with applications in antiviral defense and RNA regulation.