Abundance-based proteomics techniques do not always accurately represent the functional output, or activity, of enzymes. Following protein translation, the catalytic activity of enzymes is often further modulated through post-translational modifications or allosteric regulation, such as protein-protein interactions.
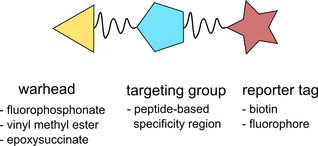
Activity-based protein profiling (ABPP) uses small molecule activity-based probes (ABPs) which form irreversible bonds with target enzymes based on their structure and their catalytic activity. ABPs consist of three distinct components: a reactive warhead to form the bond between the enzyme and the probe, a targeting group responsible for directing the probe to an enzyme’s active site, and a reporter tag. While many probes can label a large range of enzymes, increased selectivity can be achieved by tailoring the targeting group on the probe to specific enzymes of interest.
In our lab, we use ABPP to profile changes in enzyme activity during host-pathogen interactions. This allow us to identify changes in gene function due to infection which are not always abundance based. We use a variety of activity-based probes to target subsets of active enzymes and get a clear picture of the functional proteome during healthy and diseased states. This approach allows us to identify potential druggable targets for treating infections.
Select reviews:
Desrochers, G. F., Pezacki, J. P. “ABPP and Host-Virus Interactions.” Curr. Topics Microbiol. Immunol. 2019, 420:131 154, DOI:10.1007/82_2018_139. (Book Chapter)
Strmiskova M, Desrochers GF, Powdrill MH, Shaw T, Lafreniere MA, Pezacki JP “Chemical Methods for Probing Virus-Host Proteomic Interactions”ACS Infect. Dis. 2016, 2 (11), 773-786
Select research articles:
Desrochers GF, Cornacchia C, McKay CS, Pezacki JP "Activity-Based PIK Probes Detect Changes to Protein-Protein Interactions During Hepatitis C Virus Replication" ACS Infect. Dis. 2018, 4(5), 752-757
Nasheri N, Ning Z, Figeys D, Goto NK, Yao S, Pezacki JP “Activity-based profiling of the proteasome pathway during hepatitis C virus infection”, Proteomics. 2015, 15, 3815-3825.
Desrochers GF, Sherratt AR, Blais DR, Nasheri N, Ning Z, Figeys D, Goto NK Pezacki JP “Profiling protein kinase activity during hepatitis C virus replication using a Wortmannin-based probe”, ACS Inf. Dis. 2015, 1, 443-452.
Nasheri N, Joyce M, Rouleau Y, Yang P, Yao S, Tyrrell DL, Pezacki JP “Modulation of Fatty Acid Synthase Enzyme Activity and Expression during Hepatitis C Virus Replication” Chem. Biol. 2013, 20, 570-582 (Cover article).
Desrochers, G. F., Pezacki, J. P. “ABPP and Host-Virus Interactions.” Curr. Topics Microbiol. Immunol. 2019, 420:131 154, DOI:10.1007/82_2018_139. (Book Chapter)
Strmiskova M, Desrochers GF, Powdrill MH, Shaw T, Lafreniere MA, Pezacki JP “Chemical Methods for Probing Virus-Host Proteomic Interactions”ACS Infect. Dis. 2016, 2 (11), 773-786
Select research articles:
Desrochers GF, Cornacchia C, McKay CS, Pezacki JP "Activity-Based PIK Probes Detect Changes to Protein-Protein Interactions During Hepatitis C Virus Replication" ACS Infect. Dis. 2018, 4(5), 752-757
Nasheri N, Ning Z, Figeys D, Goto NK, Yao S, Pezacki JP “Activity-based profiling of the proteasome pathway during hepatitis C virus infection”, Proteomics. 2015, 15, 3815-3825.
Desrochers GF, Sherratt AR, Blais DR, Nasheri N, Ning Z, Figeys D, Goto NK Pezacki JP “Profiling protein kinase activity during hepatitis C virus replication using a Wortmannin-based probe”, ACS Inf. Dis. 2015, 1, 443-452.
Nasheri N, Joyce M, Rouleau Y, Yang P, Yao S, Tyrrell DL, Pezacki JP “Modulation of Fatty Acid Synthase Enzyme Activity and Expression during Hepatitis C Virus Replication” Chem. Biol. 2013, 20, 570-582 (Cover article).