Protein Engineering & Computational Protein Design
We specialize in the development of novel proteins with useful properties for their application in chemistry and biology. In particular, we are interested in developing designer biocatalysts for chemical synthesis as well as genetically-encoded biosensors based on fluorescent proteins for in vivo imaging applications. To achieve these goals, we develop and exploit multistate computational protein design methods to predict mutations leading to the desired protein property. More information on our research projects can be found in the Research section.
Latest Results
St-Jacques et al. (2023) Nature Communications, 14, 6058.
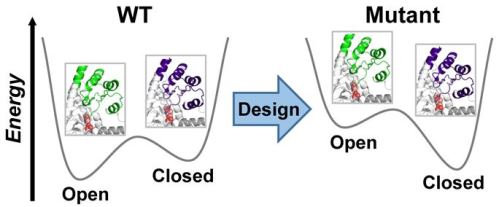
Structural plasticity of enzymes dictates their function. Yet, our ability to rationally remodel enzyme conformational landscapes to tailor catalytic properties remains limited. Here, we report a computational procedure for tuning conformational landscapes that is based on multistate design of hinge-mediated domain motions. Using this method, we redesign the conformational landscape of a natural aminotransferase to preferentially stabilize a less populated but reactive conformation and thereby increase catalytic efficiency with a non-native substrate, resulting in altered substrate selectivity. Steady-state kinetics of designed variants reveals activity increases with the non-native substrate of approximately 100-fold and selectivity switches of up to 1900-fold. Structural analyses by room-temperature X-ray crystallography and multitemperature nuclear magnetic resonance spectroscopy confirm that conformational equilibria favour the target conformation. Our computational approach opens the door to targeted alterations of conformational states and equilibria, which should facilitate the design of biocatalysts with customized activity and selectivity.
Read more here.
Previous results can be found here.
Advanced Protein Engineering Training, Internships, Courses & Exhibition (APRENTICE) Program
Interested in pursuing graduate training in protein engineering? Find out more about the APRENTICE program by clicking on the image below.
Research Funding
We gratefully acknowledge support from the following agencies:

Keywords
Protein Engineering, Computational Protein Design, Enzyme Design, Enzymes, Biocatalysis, Fluorescent Proteins, Protein Dynamics, Molecular Modeling, Protein Science, Biological Chemistry
Contact info
Roberto Chica, Ph. D. (613) 562-5800 x 1988 |
 |
|---|
