Bacterial Cell Division
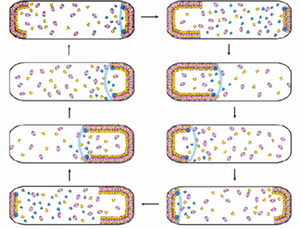
Bacterial propagation relies on precise regulation of cell division. Proper placement of the cell division septum is critical to ensure that two daughter cells of the same size can be produced with an equal amount of genetic material. A group of proteins called MinC, MinD and MinE act at the cell membrane of Gram negative bacteria to ensure that cell division occurs symmetrically. These proteins form subcellular structures on the inner membrane, with dynamic oscillation from pole to pole. (Click on the left image on the left for a schematic video of this process.) We are interested in understanding how these proteins form dynamic patterns on lipid bilayers, and how ATP hydrolysis by MinD is coupled to these patterns. We focus on the Min system from N. gonorrhoeae - ulitmately, by understanding how the Min system works we can develop new strategies to inhibit cell division and combat infection by bacterial pathogens.
Membrane Proteins:
Membrane proteins are molecular machines, essential for communication and transport across the cell membrane. In order to understand how membrane proteins can perform these functions, three-dimensional structural information is required at an atomic level of detail. New developments in solution NMR technology and methodology have made it possible for this approach to be used to determine structures for membrane proteins of increasing size and complexity.
![trp mutant struct2]3](_Media/trp_mutant_struct23_med.png)
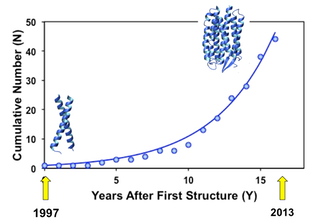
This graph shows the exponential increase in the number of unique membrane proteins for which structures have been determined by solution NMR. (The glyocphorin A dimer structure determined in 1997 (PDB ID: 1AFO) and the phototaxis receptor sensory rhodopsin II (2KSY) in 2010.)
To determine these structures the protein must be transferred from the cellular membrane into a detergent solvent that mimics its native environment. We are interested in understanding how interactions within the membrane are affected by solubilization agents used for this purpose. Regions that exist outside the membrane can also affect the structure and function of elements that reside inside the membrane. By characterizing these effects we will better understand how changes in lipid environment can alter membrane protein structure and function in biological systems. Our focus for these studies is an integral membrane protein called the rhomboid protease GlpG (x-ray structure shown on the left; PDB ID 2IC8). Members of the rhomboid family play a role in a wide range of biological processes, including parasite invasion, bacterial quorum sensing and the degradation of misfolded membrane proteins. We seek to understand how the rhomboid protease can perform these vital functions in the hydrophobic environment of the lipid membrane.